How to browse UPObase?
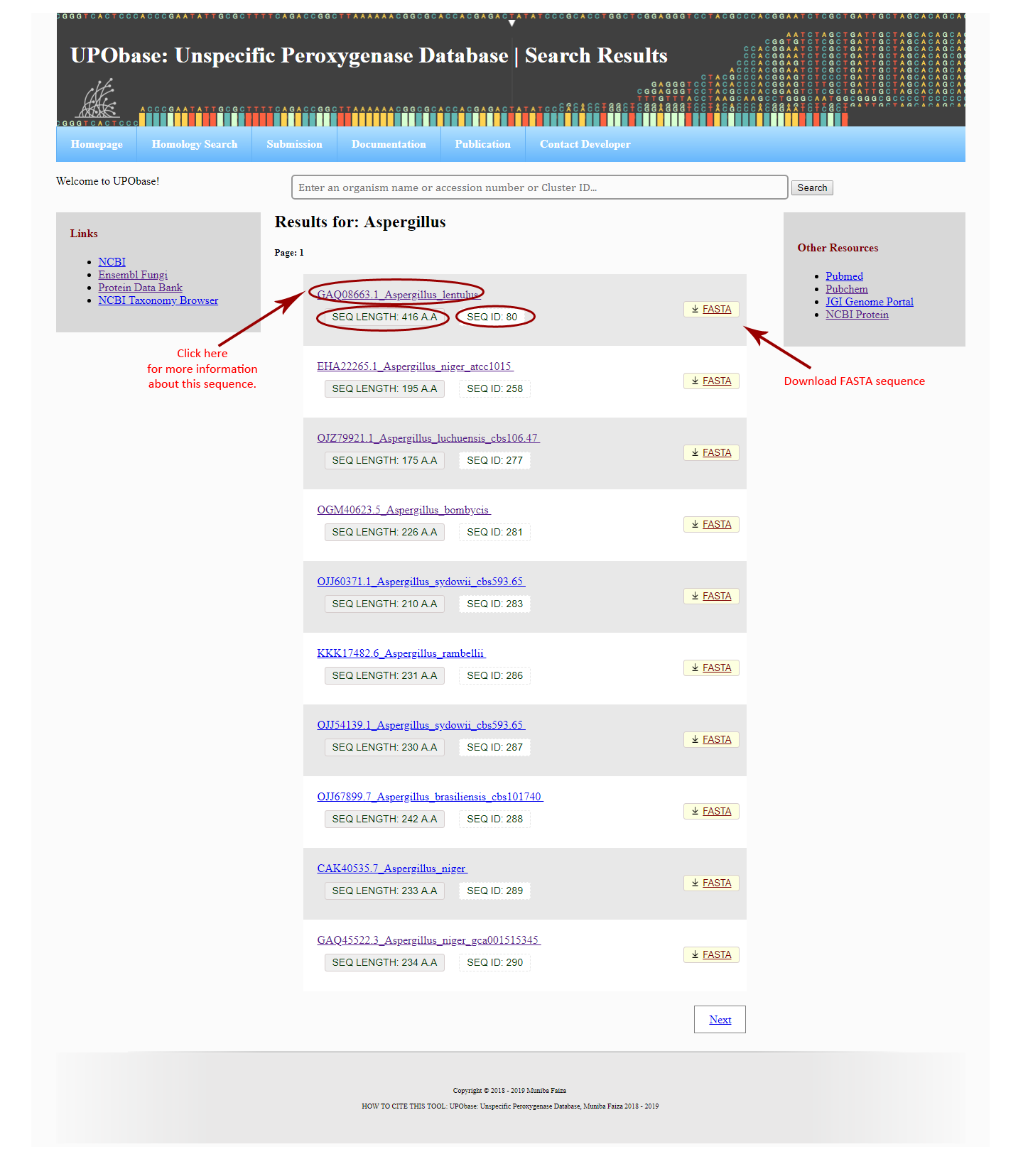
The UPObase offers to search in the database either by entering the name of an organism, an accession number, or a cluster ID in the global search box. For the ease of users, the global search box is provided on each page of the database.
In order to see the sequence details, click on any one search results displayed as a list, where all sequence information is displayed including accession number, sequence ID, cluster ID, classification, motif patterns, and detailed information of the functional roles of the motifs found in UPOs.
A FASTA sequence and homologous sequences can be downloaded from the same sequence details page. The sequence can be subjected to analyses such as alignment, phylogenetic tree, and the corresponding percent identity matrix.
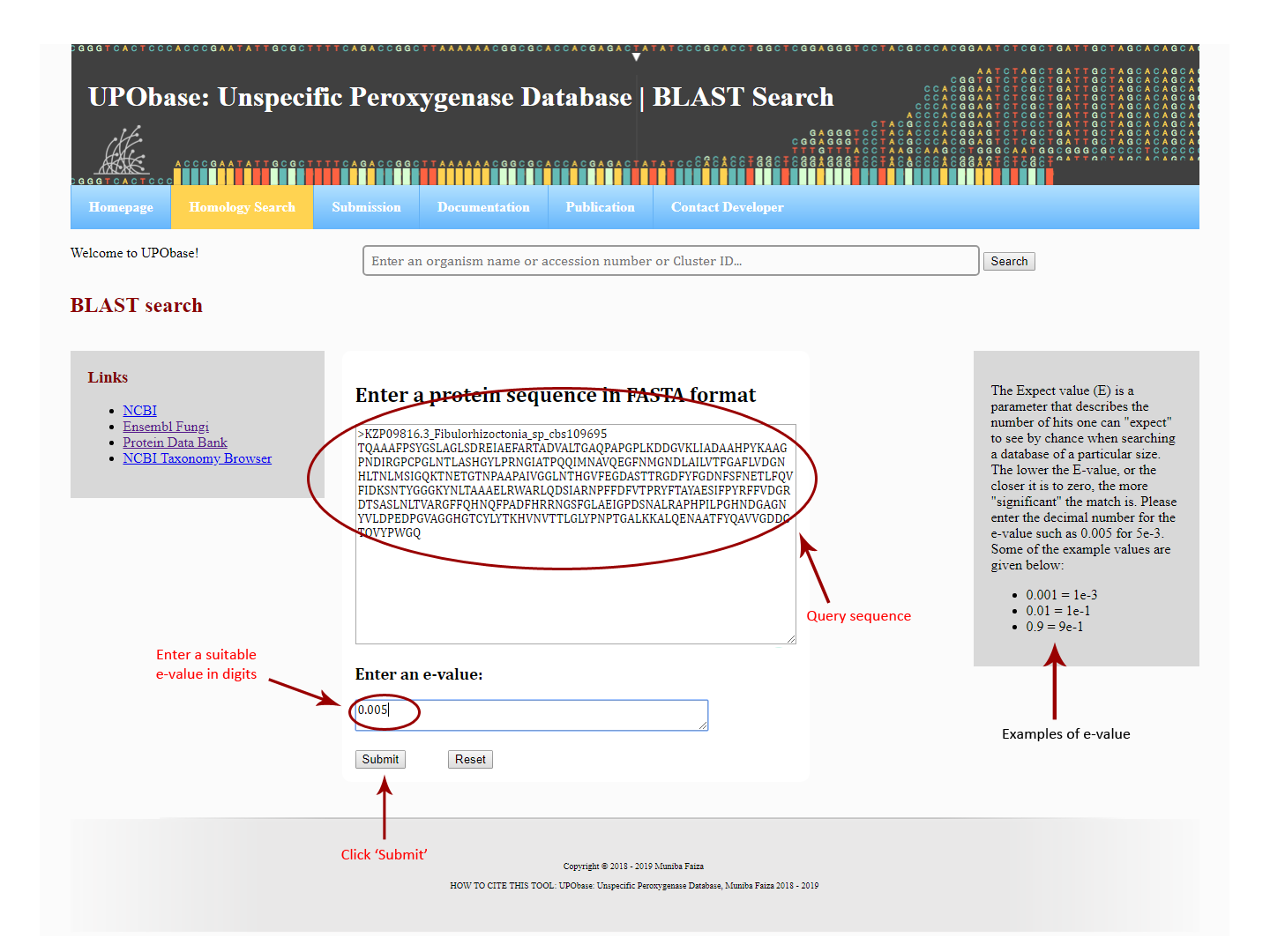
For a BLAST search against the UPObase, users have to input a protein sequence in FASTA format and an e-value in digits (e.g., 0.0005). After the job is finished, the BLAST results are displayed consisting of a list of best hits and e-value scores along with a query-subject alignment.
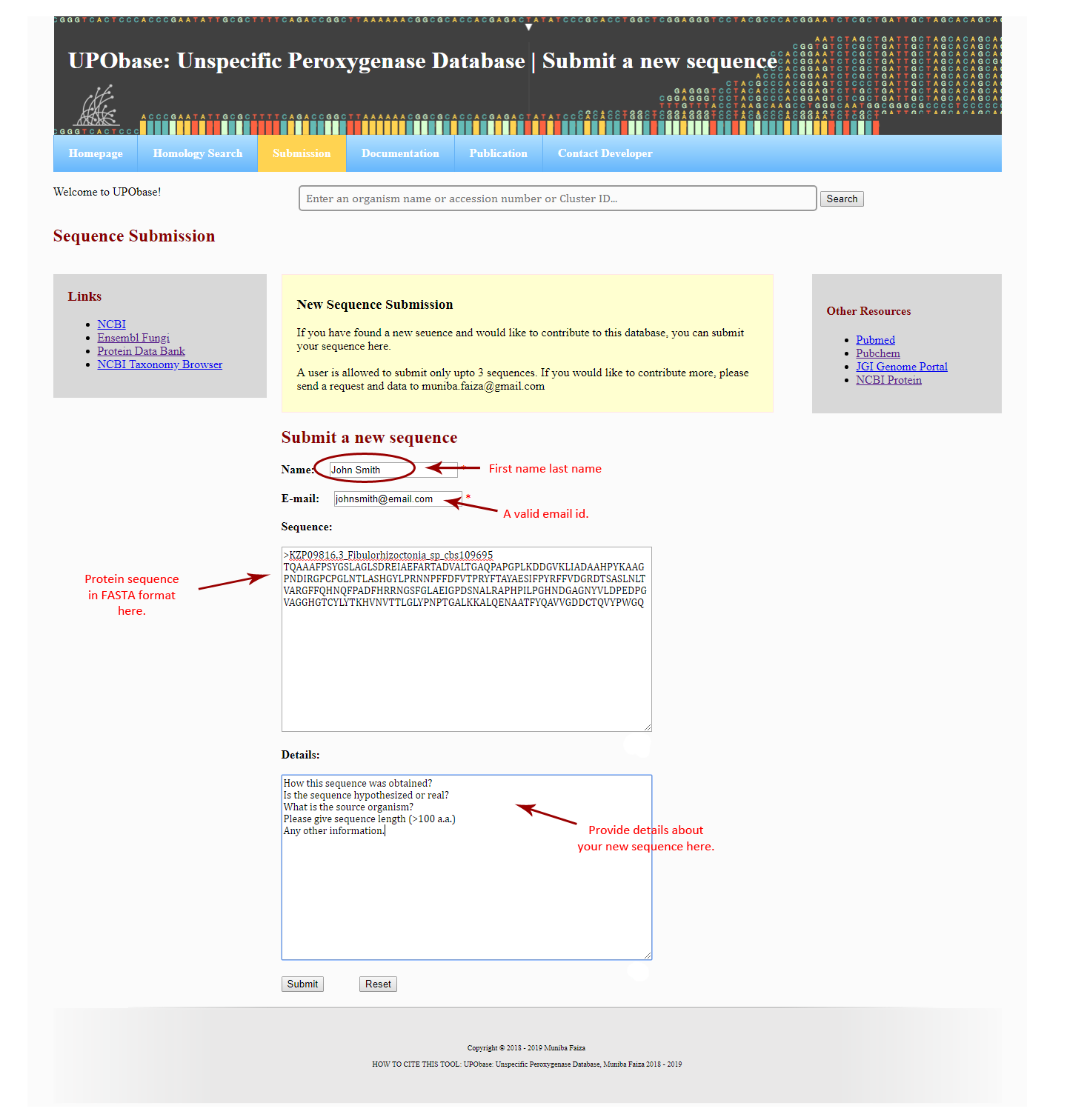
Users can also submit new sequences to UPObase via submission portal. User has to provide his name and email address (both are mandatory) and a protein sequence in FASTA format. Additionally, a user can submit only up to a maximum of three sequences.
If you have any query related to the database or face any difficulty, you can contact us on the information provided on the Contact us page.
Some examples are shown in the following section with screenshots to guide browsing of UPObase.
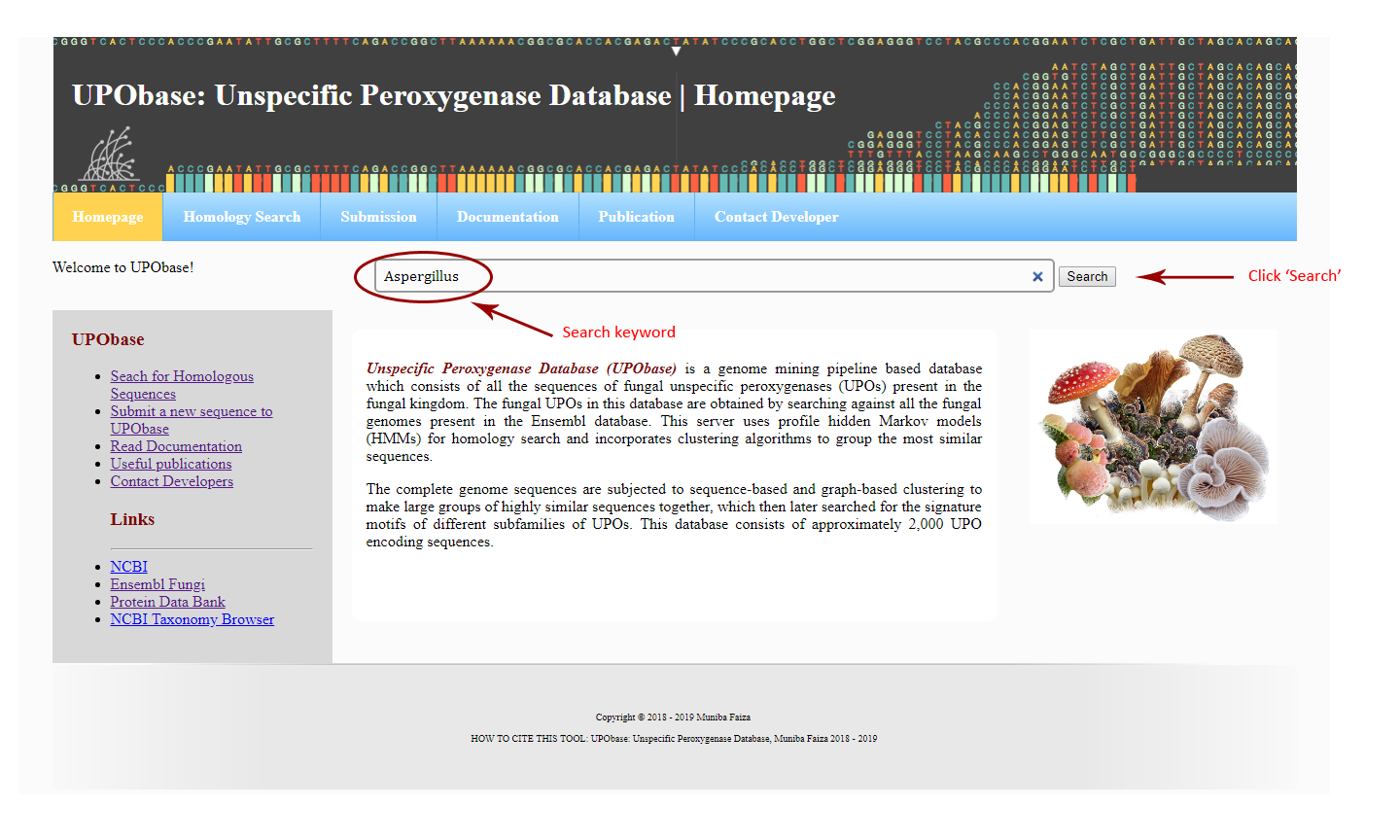
How to browse UPObase using a keyword:
You can enter a cluster ID, or accession number, or an organism name such as "Aspergillus".
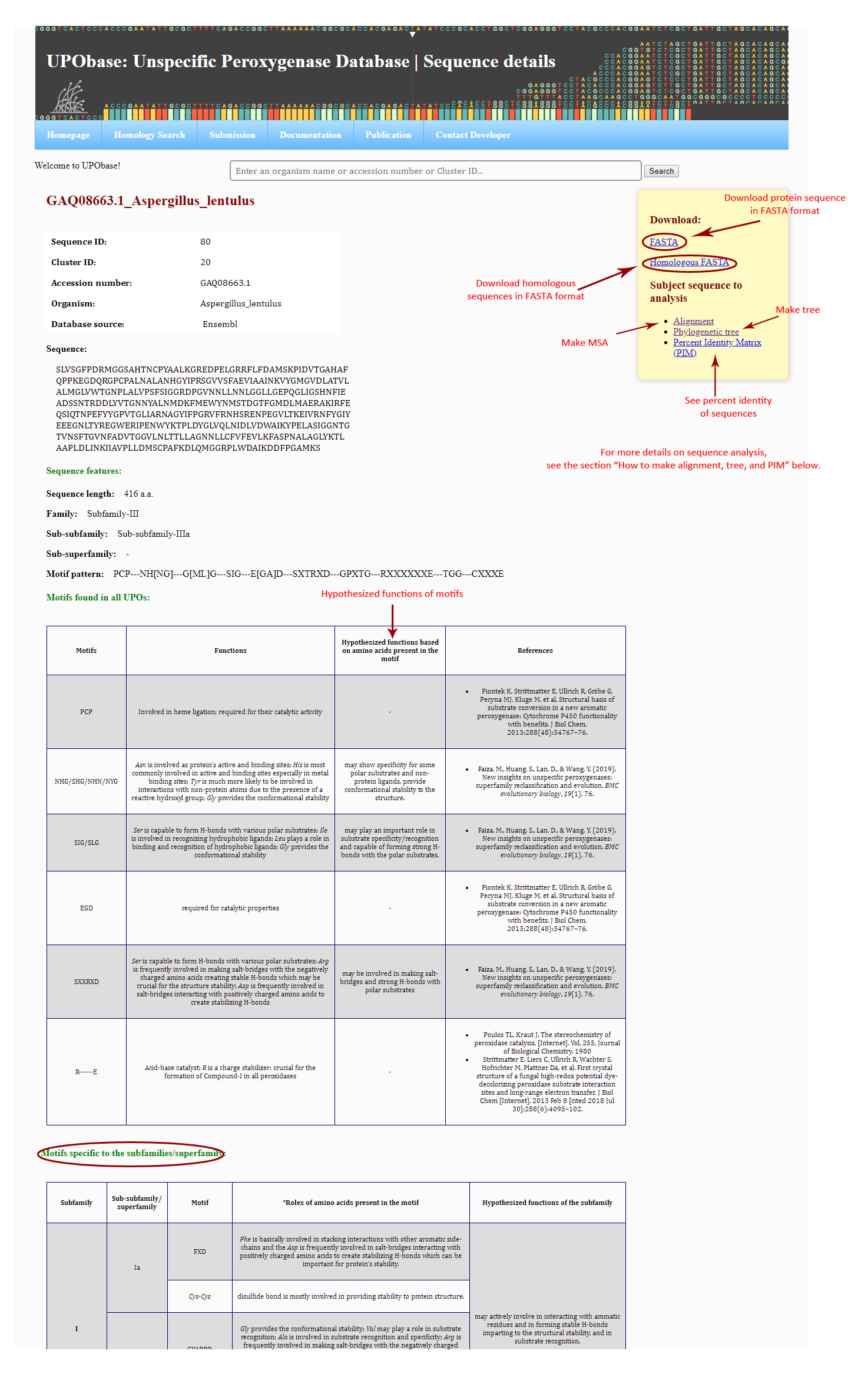
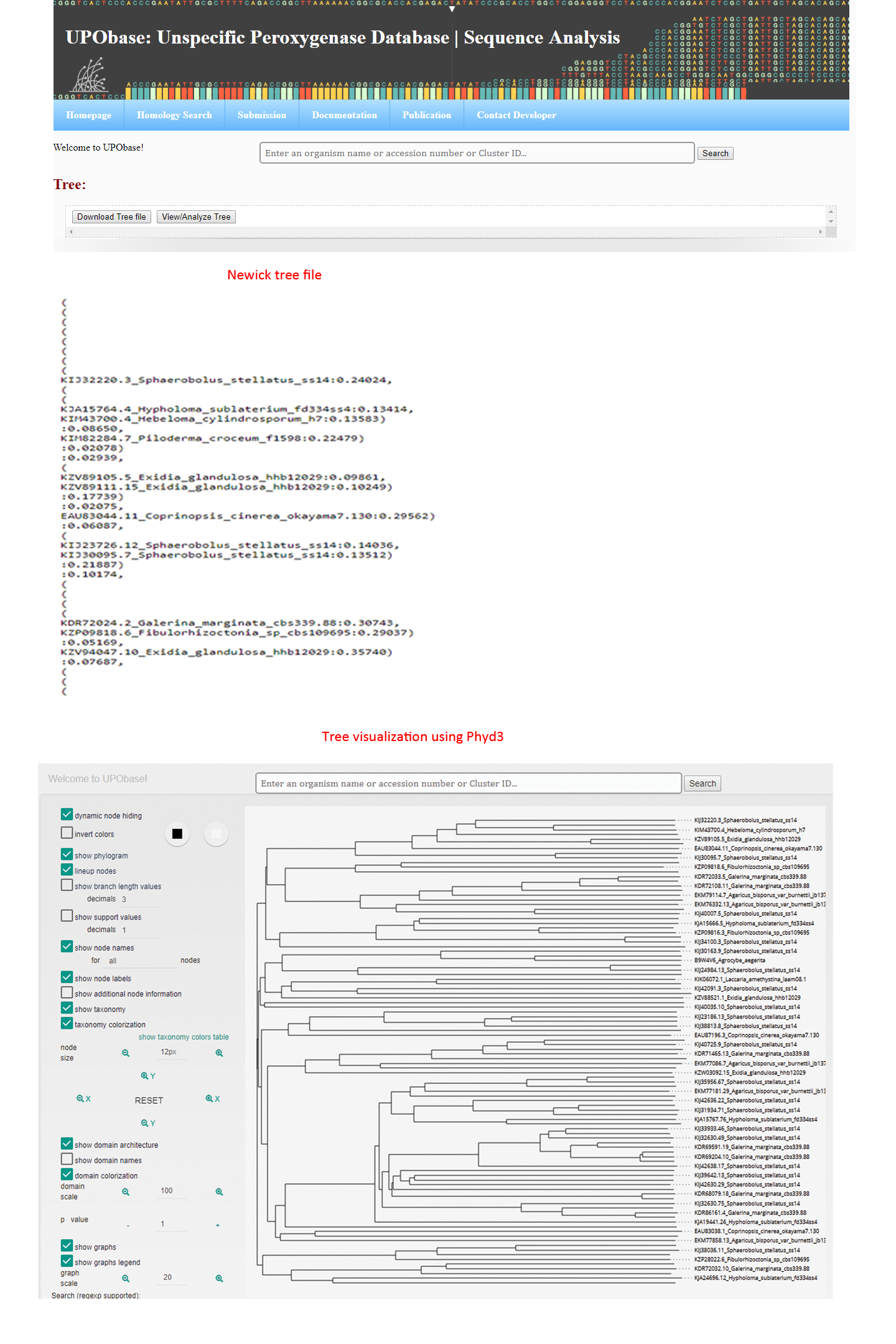
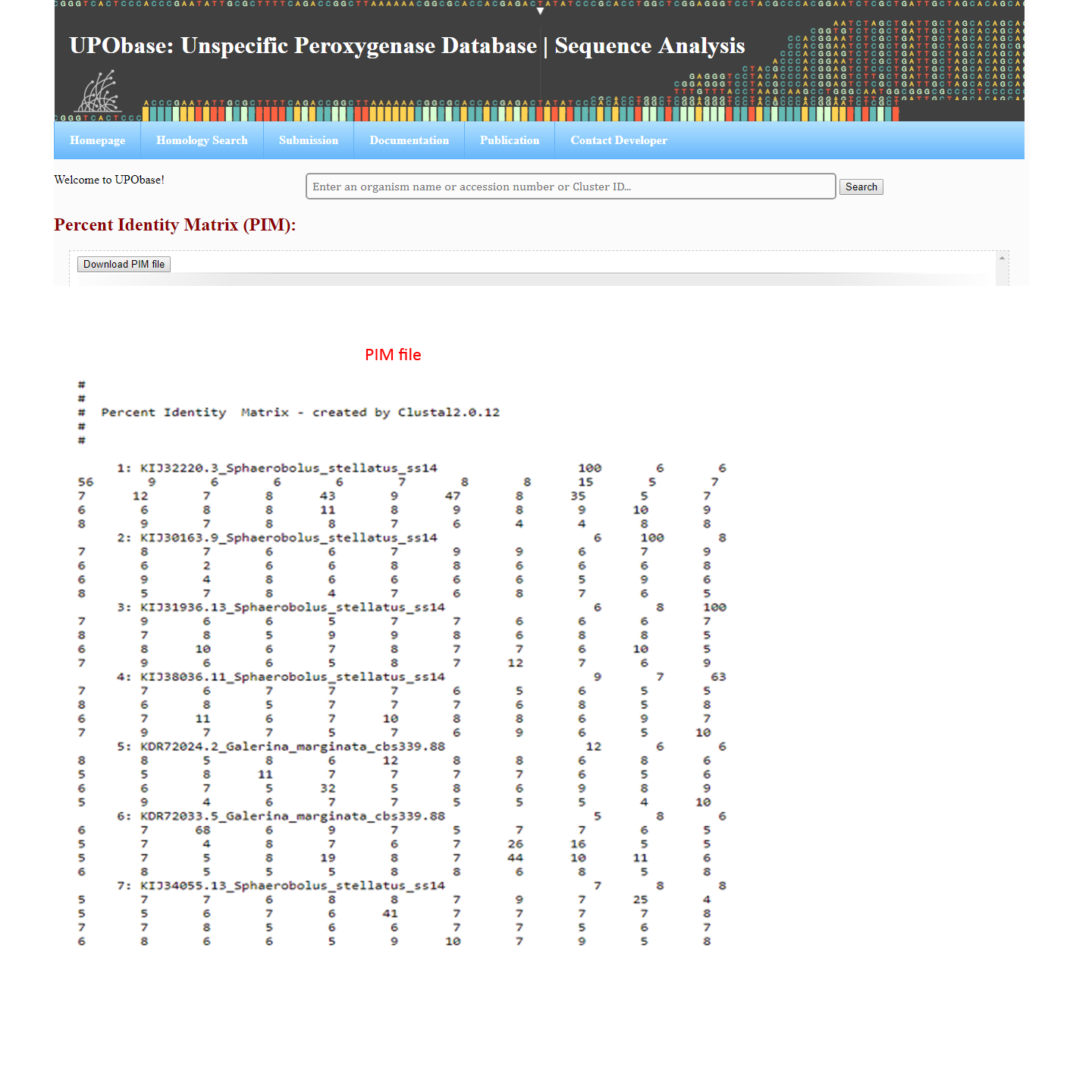
How to generate an MSA, phylogenetic tree, and percent identity matrix?
If you want to align this sequence with the other sequences in the database, then click 'Alignment', to see the corresssponding phylogenetic tree, click 'Phylogenetic tree' and for PIM, click 'Percent Identity Matrix (PIM)' as shown in the above screenshot. The result files will look like this:

How to blast against UPObase?
Enter a protein FASTA sequence and an e-value in digits as shown below. The output file will look like this.
How to submit a new sequence in UPObase?